1. mapping
1.1 CID匹配和过滤统计结果
Bash
$ cat /path/to/output/01T.mapping/V350248064_L01_read_1.CIDMap.stat
...
unique_CID_in_mask: 551164442
unique_CID_in_fq: 82769358
total_reads: 325558079
CID_with_N_reads: 223 0.00 %
CID_mapped_reads: 263145167 80.83 %
CID_exactly_mapped_reads: 215239025 66.11 %
CID_mapped_reads_with_mismatch: 47905919 14.72 %
discarded_MID_reads: 9502982 2.92 %
MID_with_N_reads: 62668 0.02 %
MID_with_polyA_reads: 7625 0.00 %
MID_with_low_quality_base_reads: 9432689 2.90 %
reads_with_dnb: 14643107 4.50 %
reads_with_adapter: 11536847 3.54 %
short_reads_filtered_with_polyA: 7402775 2.27 %
reads_with_polyA: 9930604 3.05 %
reads_with_rRNA: 0 0.00 %
Q10_bases_in_seq: 99.20 %
Q20_bases_in_seq: 97.40 %
Q30_bases_in_seq: 91.14 %
Q10_bases_in_MID: 98.29 %
Q20_bases_in_MID: 95.14 %
Q30_bases_in_MID: 85.33 %
Q10_bases_in_CID: 98.99 %
Q20_bases_in_CID: 96.99 %
Q30_bases_in_CID: 89.18 %
1.2 参考基因组比对统计
bash
$ cat /path/to/output/01T.mapping/V350248064_L01_read_1.Log.final.out
...
Number of input reads | 220059456
Average input read length | 49
UNIQUE READS:
Uniquely mapped reads number | 101482044
Uniquely mapped reads % | 46.12%
Average mapped length | 47.22
Number of splices: Total | 2446345
Number of splices: Annotated (sjdb) | 2108691
Number of splices: GT/AG | 2315419
Number of splices: GC/AG | 24032
Number of splices: AT/AC | 2288
Number of splices: Non-canonical | 104606
Mismatch rate per base, % | 1.70%
Deletion rate per base | 0.04%
Deletion average length | 2.88
Insertion rate per base | 0.01%
Insertion average length | 1.08
MULTI-MAPPING READS:
Number of reads mapped to multiple loci | 62340513
% of reads mapped to multiple loci | 28.33%
Number of reads mapped to too many loci | 3625826
% of reads mapped to too many loci | 1.65%
UNMAPPED READS:
Number of reads unmapped: too many mismatches | 0
% of reads unmapped: too many mismatches | 0.00%
Number of reads unmapped: too short | 52344265
% of reads unmapped: too short | 23.79%
Number of reads unmapped: other | 266808
% of reads unmapped: other | 0.12%
CHIMERIC READS:
Number of chimeric reads | 0
% of chimeric reads | 0.00%
1.3 mapping BAM示例
bash
$ samtools view /path/to/output/01T.mapping/V350248064_L01_read_1.Aligned.sortedByCoord.out.bam | head -2
V350248064L1C002R00801053349 0 1 3000093 255 13S37M * 0 0 TTAGACCACTTGATGTGCTTTTTTTTTTTTTTTTTTTTTTTTTTGGGTGG BCBA>FFE96CB7;C-;48A<9@BCC@C;BADBABDCDAD@C5.+#4-@@ NH:i:1 HI:i:1 AS:i:34 nM:i:1 Cx:i:18567 Cy:i:12717 UR:Z:C559D
V350248064L1C003R04700229281 0 1 3000093 3 13S36M1S * 0 0 ACTTAGCACTTGATGTGCTTTTTTTTTTTTTTTTTTTTTTTTTTGGGTGT B@DDECDDE?=E0?C;>??C@DAADDCBDDDCDDCCBBCCDDD)''&#"BNH:i:2 HI:i:1 AS:i:33 nM:i:1 Cx:i:14384 Cy:i:8392 UR:Z:72BC7
2. merge
2.1 CID对应reads数列表示例
bash
$ head /path/to/output/02T.merge/A02677B5.merge.barcodeReadsCount.txt
0 4885 1
0 5494 1
0 7252 1
0 7263 1
0 7392 1
0 8929 1
0 9063 2
0 12673 7
0 12845 4
0 14828 1
3. count
3.1 MID过滤和基因注释结果统计
bash
$ cat /path/to/output/03T.count/A02677B5.Aligned.sortedByCoord.out.merge.q10.dedup.target.bam.summary.stat
## FILTER & DEDUPLICATION METRICS
TOTAL_READS PASS_FILTER ANNOTATED_READS UNIQUE_READS FAIL_FILTER_RATE FAIL_ANNOTATE_RATEDUPLICATION_RATE
497134569 306509812 203236286 112788155 38.34 33.69 44.50
## ANNOTATION METRICS
TOTAL_READS MAP EXONIC INTRONIC INTERGENIC TRANSCRIPTOME ANTISENSE
306509812 306509812 159125426 44110860 103273526 203236286 89162163
100.0 100.0 51.9 14.4 33.7 66.3 29.1
3.2 注释结果BAM示例
$ samtools view /path/to/output/03T.count/A02677B5.Aligned.sortedByCoord.out.merge.q10.dedup.target.bam | head -2
V350248064L1C002R00801053349 0 1 3000093 255 13S37M * 0 0 TTAGACCACTTGATGTGCTTTTTTTTTTTTTTTTTTTTTTTTTTGGGTGG BCBA>FFE96CB7;C-;48A<9@BCC@C;BADBABDCDAD@C5.+#4-@@ NH:i:1 HI:i:1 AS:i:34 nM:i:1 Cx:i:18567 Cy:i:12717 UR:Z:C559D XF:i:2
V350248064L1C003R04700229281 512 1 3000093 3 13S36M1S * 0 0 ACTTAGCACTTGATGTGCTTTTTTTTTTTTTTTTTTTTTTTTTTGGGTGT B@DDECDDE?=E0?C;>??C@DAADDCBDDDCDDCCBBCCDDD)''&#"BNH:i:2 HI:i:1 AS:i:33 nM:i:1 Cx:i:14384 Cy:i:8392 UR:Z:72BC7
3.3 count基因表达文件示例
bash
$ h5dump -n /path/to/output/03T.count/A02677B5.raw.gef
HDF5 "/path/to/output/03T.count/A02677B5.raw.gef" {
FILE_CONTENTS {
group /
group /geneExp
group /geneExp/bin1
dataset /geneExp/bin1/exon
dataset /geneExp/bin1/expression
dataset /geneExp/bin1/gene
}
}
$ h5dump -d /geneExp/bin1/expression /path/to/output/03T.count/A02677B5.raw.gef | head -15
HDF5 "/path/to/output/03T.count/A02677B5.raw.gef" {
DATASET "/geneExp/bin1/expression" {
DATATYPE H5T_COMPOUND {
H5T_STD_U32LE "x";
H5T_STD_U32LE "y";
H5T_STD_U8LE "count";
}
DATASPACE SIMPLE { ( 97081672 ) / ( 97081672 ) }
DATA {
(0): {
2630,
8744,
1
},
(1): {
$ h5dump -d /geneExp/bin1/gene /path/to/output/03T.count/A02677B5.raw.gef | head -20
HDF5 "/path/to/output/03T.count/A02677B5.raw.gef" {
DATASET "/geneExp/bin1/gene" {
DATATYPE H5T_COMPOUND {
H5T_STRING {
STRSIZE 64;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
} "gene";
H5T_STD_U32LE "offset";
H5T_STD_U32LE "count";
}
DATASPACE SIMPLE { ( 27445 ) / ( 27445 ) }
DATA {
(0): {
"0610005C13Rik",
0,
799
},
(1): {
3.4 count抽样文件
bash
$ head -8 /path/to/output/03T.count/A02677B5_raw_barcode_gene_exp.txt
x y geneIndex MIDIndex readCount
12053 13111 14468 282639 2
7295 9953 14469 599164 1
21748 23094 14469 477127 1
11735 13821 14470 400133 5
16697 12775 14470 411141 1
3790 22503 14470 119157 1
13954 13219 14470 413356 1
4. mapping-SP
4.1 CID 比对、MID 过滤 和 PID 比对统计
bash
$ cat /path/to/01P.mapping/A02677B5_map.stat
...
unique_CID_in_mask: 551164442
unique_CID_in_fq: 74809591
total_reads: 573659235
duplication_rate: 59.07 %
CID_mapped_reads: 475103289 82.82 %
CID_exactly_mapped_read: 404140856 70.45 %
CID_mapped_reads_with_mismatch: 70962433 12.37 %
CID_with_N_reads: 7354 0.00 %
discarded_MID_reads: 1489710 0.26 %
MID_with_N_reads: 4583 0.00 %
MID_with_low_quality_base_reads: 1485127 0.26 %
PID_mapped_reads: 427761398 90.32 %
PID_exactly_mapped_reads: 398063274 84.05 %
PID_mapped_reads_with_mismatch: 29698124 6.27 %
PID_mapped_unique_reads: 175086793 36.97 %
Q10_bases_in_CID: 99.70 %
Q20_bases_in_CID: 97.63 %
Q30_bases_in_CID: 87.72 %
Q10_bases_in_MID: 99.63 %
Q20_bases_in_MID: 97.41 %
Q30_bases_in_MID: 88.10 %
Q10_bases_in_ADT: 99.49 %
Q20_bases_in_ADT: 97.49 %
Q30_bases_in_ADT: 93.44 %
4.2 CID 对应reads数列表示例
bash
$ head /path/to/01P.mapping/A02677B5_valid_cid_reads.tsv
x y readCount
13012 22382 2
4261 13080 2
16225 13353 14
9533 4088 4
18486 23899 3
13971 16291 9
9013 5905 1
4549 8517 3
13751 18063 23
4.3 mapping-SP 抽样文件示例
bash
$ head /path/to/01P.mapping/A02677B5_cid_pid_mid_reads.tsv
x y PIDIndex MID readCount
232 26091 0 291549 4
2042 8307 0 611198 3
2238 3469 0 483548 1
2525 1134 0 417813 3
2616 20068 0 788601 7
3557 11877 0 416037 2
3698 10004 0 419111 1
3698 19417 0 422891 1
3698 19547 0 139153 1
4.4 mapping-SP蛋白表达矩阵文件示例
$ h5dump -n /path/to/01P.mapping-SP/A02677B5.protein.raw.gef
HDF5 "/path/to/01P.mapping-SP/A02677B5.protein.raw.gef" {
FILE_CONTENTS {
group /
group /geneExp
group /geneExp/bin1
dataset /geneExp/bin1/expression
dataset /geneExp/bin1/gene
}
}
$ h5dump -d /geneExp/bin1/expression /path/to/01P.mapping-SP/A02677B5.protein.raw.gef|head -15
HDF5 "/path/to/01P.mapping-SP/A02677B5.protein.raw.gef" {
DATASET "/geneExp/bin1/expression" {
DATATYPE H5T_COMPOUND {
H5T_STD_I32LE "x";
H5T_STD_I32LE "y";
H5T_STD_U8LE "count";
}
DATASPACE SIMPLE { ( 152263461 ) / ( 152263461 ) }
DATA {
(0): {
11408,
5590,
2
},
(1): {
$ h5dump -d /geneExp/bin1/gene /path/to/01P.mapping-SP/A02677B5.protein.raw.gef|head -20
HDF5 "/path/to/01P.mapping-SP/A02677B5.protein.raw.gef" {
DATASET "/geneExp/bin1/gene" {
DATATYPE H5T_COMPOUND {
H5T_STRING {
STRSIZE 64;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
} "gene";
H5T_STD_U32LE "offset";
H5T_STD_U32LE "count";
}
DATASPACE SIMPLE { ( 128 ) / ( 128 ) }
DATA {
(0): {
"Ly49D_Ms",
0,
45274
},
(1): {
5. calibration
5.1 校准后的完整GEFs 基因表达矩阵示例
bash
$ h5dump -n /path/to/output/04.calibration/A02677B5.gef
HDF5 "/path/to/output/04.calibration/A02677B5.gef" {
FILE_CONTENTS {
group /
group /geneExp
group /geneExp/bin1
dataset /geneExp/bin1/exon
dataset /geneExp/bin1/expression
dataset /geneExp/bin1/gene
group /geneExp/bin10
dataset /geneExp/bin10/exon
dataset /geneExp/bin10/expression
dataset /geneExp/bin10/gene
group /geneExp/bin100
dataset /geneExp/bin100/exon
dataset /geneExp/bin100/expression
dataset /geneExp/bin100/gene
group /geneExp/bin150
dataset /geneExp/bin150/exon
dataset /geneExp/bin150/expression
dataset /geneExp/bin150/gene
group /geneExp/bin20
dataset /geneExp/bin20/exon
dataset /geneExp/bin20/expression
dataset /geneExp/bin20/gene
group /geneExp/bin200
dataset /geneExp/bin200/exon
dataset /geneExp/bin200/expression
dataset /geneExp/bin200/gene
group /geneExp/bin5
dataset /geneExp/bin5/exon
dataset /geneExp/bin5/expression
dataset /geneExp/bin5/gene
group /geneExp/bin50
dataset /geneExp/bin50/exon
dataset /geneExp/bin50/expression
dataset /geneExp/bin50/gene
group /stat
dataset /stat/gene
group /wholeExp
dataset /wholeExp/bin1
dataset /wholeExp/bin10
dataset /wholeExp/bin100
dataset /wholeExp/bin150
dataset /wholeExp/bin20
dataset /wholeExp/bin200
dataset /wholeExp/bin5
dataset /wholeExp/bin50
group /wholeExpExon
dataset /wholeExpExon/bin1
dataset /wholeExpExon/bin10
dataset /wholeExpExon/bin100
dataset /wholeExpExon/bin150
dataset /wholeExpExon/bin20
dataset /wholeExpExon/bin200
dataset /wholeExpExon/bin5
dataset /wholeExpExon/bin50
}
}
$ h5dump -d /stat/gene /path/to/output/04.calibration/A02677B5.gef | head -20
HDF5 "/path/to/output/04.calibration/A02677B5.gef" {
DATASET "/stat/gene" {
DATATYPE H5T_COMPOUND {
H5T_STRING {
STRSIZE 64;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
} "gene";
H5T_STD_U32LE "MIDcount";
H5T_IEEE_F32LE "E10";
}
DATASPACE SIMPLE { ( 27445 ) / ( 27445 ) }
DATA {
(0): {
"Gm42418",
24328473,
77.1551
},
(1): {
$ h5dump -n /path/to/output/04.calibration/A02677B5.protein.gef
HDF5 "/path/to/output/04.calibration/A02677B5.protein.gef" {
FILE_CONTENTS {
group /
group /geneExp
group /geneExp/bin1
dataset /geneExp/bin1/expression
dataset /geneExp/bin1/gene
group /geneExp/bin10
dataset /geneExp/bin10/expression
dataset /geneExp/bin10/gene
group /geneExp/bin100
dataset /geneExp/bin100/expression
dataset /geneExp/bin100/gene
group /geneExp/bin150
dataset /geneExp/bin150/expression
dataset /geneExp/bin150/gene
group /geneExp/bin20
dataset /geneExp/bin20/expression
dataset /geneExp/bin20/gene
group /geneExp/bin200
dataset /geneExp/bin200/expression
dataset /geneExp/bin200/gene
group /geneExp/bin5
dataset /geneExp/bin5/expression
dataset /geneExp/bin5/gene
group /geneExp/bin50
dataset /geneExp/bin50/expression
dataset /geneExp/bin50/gene
group /stat
dataset /stat/gene
group /wholeExp
dataset /wholeExp/bin1
dataset /wholeExp/bin10
dataset /wholeExp/bin100
dataset /wholeExp/bin150
dataset /wholeExp/bin20
dataset /wholeExp/bin200
dataset /wholeExp/bin5
dataset /wholeExp/bin50
}
}
$ h5dump -d /stat/gene /path/to/output/04.calibration/A02677B5.protein.gef | head -20
HDF5 "/path/to/output/04.calibration/A02677B5.protein.gef" {
DATASET "/stat/gene" {
DATATYPE H5T_COMPOUND {
H5T_STRING {
STRSIZE 64;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
} "gene";
H5T_STD_U32LE "MIDcount";
H5T_IEEE_F32LE "E10";
}
DATASPACE SIMPLE { ( 128 ) / ( 128 ) }
DATA {
(0): {
"CD3_Ms",
19264971,
73.0514
},
(1): {
6. register and imageTools
6.1 配准图
文件: /path/to/output/05.register/DAPI_fov_stitched_transformed.tif and /path/to/output/05.register/DAPI_A02677B5_regist.tif
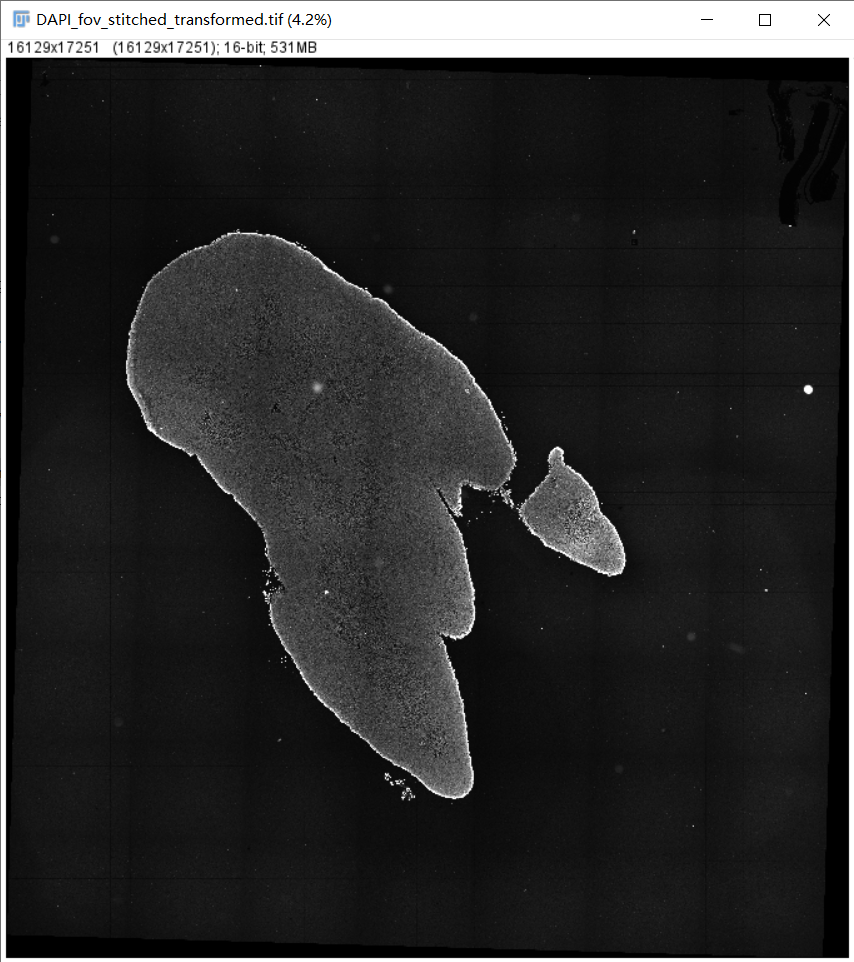
/path/to/output/05.register/DAPI_fov_stitched_transformed.tif
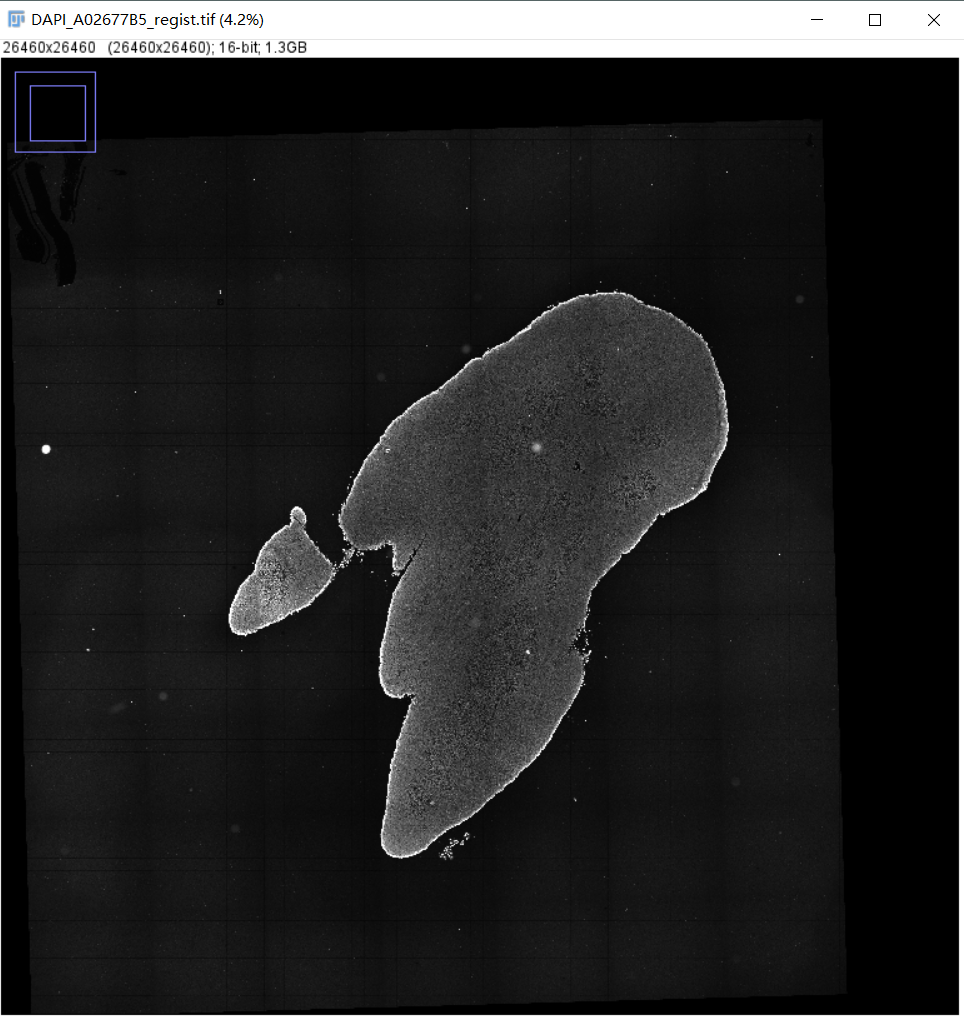
/path/to/output/05.register/DAPI_A02677B5_regist.tif
6.2 ImageTools merge
可通过ImageTools merge 确认组织和细胞分割。请查看<转录组测试数据结果展示> 4.3 ImageTools merge 部分了解更多信息。
6.3 ImageTools overlay
可通过ImageTools merge 确认组织和细胞分割。请查看<转录组测试数据结果展示> 4.4 ImageTools overlay 部分了解更多信息。
6.4 图像处理过程记录文件
bash
$ h5dump -n /path/to/output/05.register/A02677B5_SC_20240222_150321_3.0.3.ipr
HDF5 "/path/to/output/05.register/A02677B5_SC_20240222_150321_3.0.3.ipr" {
FILE_CONTENTS {
group /
group /DAPI
group /DAPI/CellSeg
dataset /DAPI/CellSeg/CellMask
dataset /DAPI/CellSeg/CellSegTrace
group /DAPI/ImageInfo
dataset /DAPI/ImageInfo/RGBScale
group /DAPI/QCInfo
dataset /DAPI/QCInfo/ClarityArr
group /DAPI/QCInfo/CrossPoints
dataset /DAPI/QCInfo/CrossPoints/0_0
...
dataset /DAPI/QCInfo/CrossPoints/8_6
group /DAPI/Register
dataset /DAPI/Register/MatrixTemplate
group /DAPI/Stitch
group /DAPI/Stitch/ScopeStitch
dataset /DAPI/Stitch/ScopeStitch/GlobalLoc
dataset /DAPI/Stitch/ScopeStitch/ScopeHorizontalJitter
dataset /DAPI/Stitch/ScopeStitch/ScopeJitterDiff
dataset /DAPI/Stitch/ScopeStitch/ScopeVerticalJitter
group /DAPI/Stitch/StitchEval
dataset /DAPI/Stitch/StitchEval/GlobalDeviation
dataset /DAPI/Stitch/StitchEval/StitchEvalH
dataset /DAPI/Stitch/StitchEval/StitchEvalV
dataset /DAPI/Stitch/TemplatePoint
dataset /DAPI/Stitch/TransformTemplate
group /DAPI/TissueSeg
dataset /DAPI/TissueSeg/TissueMask
group /ManualState
dataset /Preview
group /StereoResepSwitch
}
}
$ h5dump -A /path/to/output/05.register/A02677B5_SC_20240222_150321_3.0.3.ipr | head -20
HDF5 "/path/to/output/05.register/A02677B5_SC_20240222_150321_3.0.3.ipr" {
GROUP "/" {
ATTRIBUTE "IPRVersion" {
DATATYPE H5T_STRING {
STRSIZE H5T_VARIABLE;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_UTF8;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "0.2.0"
}
}
GROUP "DAPI" {
GROUP "CellSeg" {
ATTRIBUTE "CellSegShape" {
DATATYPE H5T_STD_I64LE
DATASPACE SIMPLE { ( 2 ) / ( 2 ) }
DATA {
7. tissueCut
7.1 组织覆盖区域统计分析
bash
$ cat /path/to/output/06T.tissuecut/tissuecut.statContour_area: 48203139
Contour_area: 48203139
Number_of_DNB_under_tissue: 23455424
Ratio: 48.66%
Total_gene_type: 26611
MID_counts: 84007203
Fraction_MID_in_spots_under_tissue: 74.48%
Reads_under_tissue: 491750982
Fraction_reads_in_spots_under_tissue: 60.89%
binSize=1
Mean_reads_per_spot: 18.28
Median_reads_per_spot: 16.00
Mean_gene_type_per_spot: 3.02
Median_gene_type_per_spot: 3
Mean_Umi_per_spot: 3.58
Median_Umi_per_spot: 3
binSize=20
Mean_reads_per_spot: 4037.53
Median_reads_per_spot: 4161.00
Mean_gene_type_per_spot: 401.42
Median_gene_type_per_spot: 415
Mean_Umi_per_spot: 689.86
Median_Umi_per_spot: 716
binSize=50
Mean_reads_per_spot: 24815.86
Median_reads_per_spot: 25896.00
Mean_gene_type_per_spot: 1868.18
Median_gene_type_per_spot: 1963
Mean_Umi_per_spot: 4239.79
Median_Umi_per_spot: 4450
binSize=100
Mean_reads_per_spot: 96725.21
Median_reads_per_spot: 102585.00
Mean_gene_type_per_spot: 4617.22
Median_gene_type_per_spot: 4912
Mean_Umi_per_spot: 16530.34
Median_Umi_per_spot: 17600
binSize=150
Mean_reads_per_spot: 211870.31
Median_reads_per_spot: 227950.00
Mean_gene_type_per_spot: 6723.55
Median_gene_type_per_spot: 7212
Mean_Umi_per_spot: 36194.40
Median_Umi_per_spot: 38985
binSize=200
Mean_reads_per_spot: 368628.94
Median_reads_per_spot: 401073.00
Mean_gene_type_per_spot: 8267.19
Median_gene_type_per_spot: 8889
Mean_Umi_per_spot: 63021.16
Median_Umi_per_spot: 68799
$ cat /path/to/06P.tissuecut/tissuecut.stat
# Tissue Statistic Analysis with Stain Image
Contour_area: 48203139
Number_of_DNB_under_tissue: 24749871
Ratio: 51.34%
Total_protein_type: 128
MID_counts: 113480216
Fraction_MID_in_spots_under_tissue: 64.84%
Reads_under_tissue: 310743393
Fraction_reads_in_spots_under_tissue: 65.41%
binSize=1
Mean_reads_per_spot: 12.42
Median_reads_per_spot: 10.00
Mean_protein_type_per_spot: 3.85
Median_protein_type_per_spot: 3
Mean_Umi_per_spot: 4.59
Median_Umi_per_spot: 4
binSize=20
Mean_reads_per_spot: 2551.64
Median_reads_per_spot: 2541.00
Mean_protein_type_per_spot: 91.29
Median_protein_type_per_spot: 93
Mean_Umi_per_spot: 931.85
Median_Umi_per_spot: 930
binSize=50
Mean_reads_per_spot: 15683.81
Median_reads_per_spot: 15822.00
Mean_protein_type_per_spot: 119.51
Median_protein_type_per_spot: 122
Mean_Umi_per_spot: 5727.85
Median_Umi_per_spot: 5791
binSize=100Mean_reads_per_spot: 61133.86
Median_reads_per_spot: 62505.00
Mean_protein_type_per_spot: 125.57
Median_protein_type_per_spot: 128
Mean_Umi_per_spot: 22325.44
Median_Umi_per_spot: 22867
binSize=150
Mean_reads_per_spot: 133883.41
Median_reads_per_spot: 139390.00
Mean_protein_type_per_spot: 125.91
Median_protein_type_per_spot: 128
Mean_Umi_per_spot: 48892.81
Median_Umi_per_spot: 51058
binSize=200
Mean_reads_per_spot: 233115.83
Median_reads_per_spot: 245086.00
Mean_protein_type_per_spot: 126.11
Median_protein_type_per_spot: 128
Mean_Umi_per_spot: 85131.45
Median_Umi_per_spot: 89608
7.2 组织覆盖区域基因表达矩阵示例
$ h5dump -n /path/to/output/06T.tissuecut/A02677B5.tissue.gef
HDF5 "/path/to/output/06T.tissuecut/A02677B5.tissue.gef" {
FILE_CONTENTS {
group /
group /geneExp
group /geneExp/bin1
dataset /geneExp/bin1/exon
dataset /geneExp/bin1/expression
dataset /geneExp/bin1/gene
}
}
$ h5dump -d /geneExp/bin1/expression /path/to/output/06T.tissuecut/A02677B5.tissue.gef | head -15
HDF5 "/path/to/output/06T.tissuecut/A02677B5.tissue.gef" {
DATASET "/geneExp/bin1/expression" {
DATATYPE H5T_COMPOUND {
H5T_STD_I32LE "x";
H5T_STD_I32LE "y";
H5T_STD_U8LE "count";
}
DATASPACE SIMPLE { ( 70724787 ) / ( 70724787 ) }
DATA {
(0): {
9713,
15266,
1
},
(1): {
$ h5dump -d /geneExp/bin1/gene /path/to/output/06T.tissuecut/A02677B5.tissue.gef | head -20
HDF5 "/path/to/output/06T.tissuecut/A02677B5.tissue.gef" {
DATASET "/geneExp/bin1/gene" {
DATATYPE H5T_COMPOUND {
H5T_STRING {
STRSIZE 64;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
} "gene";
H5T_STD_U32LE "offset";
H5T_STD_U32LE "count";
}
DATASPACE SIMPLE { ( 26611 ) / ( 26611 ) }
DATA {
(0): {
"0610005C13Rik",
0,
322
},
(1): {
$ h5dump -n /path/to/06P.tissuecut/A02677B5.protein.tissue.gef
HDF5 "/path/to/06P.tissuecut/A02677B5.protein.tissue.gef" {
FILE_CONTENTS {
group /
group /geneExp
group /geneExp/bin1
dataset /geneExp/bin1/expression
dataset /geneExp/bin1/gene
}
}$ h5dump -d /geneExp/bin1/expression /path/to/output/06P.tissuecut/A02677B5.protein.tissue.gef | head
-15HDF5 "/path/to/06P.tissuecut/A02677B5.protein.tissue.gef" {
DATASET "/geneExp/bin1/expression" {
DATATYPE H5T_COMPOUND {
H5T_STD_I32LE "x";
H5T_STD_I32LE "y";
H5T_STD_U8LE "count"; }
DATASPACE SIMPLE { ( 95273207 ) / ( 95273207 ) }
DATA {
(0): {
11411,
14924,
1
},
(1): {
$ h5dump -d /geneExp/bin1/gene /path/to/output/06P.tissuecut/A02677B5.protein.tissue.gef |
head -20HDF5 "/path/to/output/06P.tissuecut/A02677B5.protein.tissue.gef" {
DATASET "/geneExp/bin1/gene" {
DATATYPE H5T_COMPOUND {
H5T_STRING {
STRSIZE 64;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
} "gene";
H5T_STD_U32LE "offset";
H5T_STD_U32LE "count";
}
DATASPACE SIMPLE { ( 128 ) / ( 128 ) }
DATA {
(0): {
"Ly49D_Ms",
0,
16828
},
(1): {
8. cellCut
8.1 cell bin 基因表达矩阵示例
bash
$ h5dump -n /path/to/output/061T.cellcut/A02677B5.cellbin.gef
HDF5 "/path/to/output/061T.cellcut/A02677B5.cellbin.gef" {
FILE_CONTENTS {
group /
group /cellBin
dataset /cellBin/blockIndex
dataset /cellBin/blockSize
dataset /cellBin/cell
dataset /cellBin/cellBorder
dataset /cellBin/cellExon
dataset /cellBin/cellExp
dataset /cellBin/cellExpExon
dataset /cellBin/cellTypeList
dataset /cellBin/gene
dataset /cellBin/geneExon
dataset /cellBin/geneExp
dataset /cellBin/geneExpExon
}
}
$ h5dump -n /path/to/061P.cellcut/A02677B5.protein.cellbin.gef
HDF5 "/path/to/061P.cellcut/A02677B5.protein.cellbin.gef" {
FILE_CONTENTS {
group /
group /cellBin
dataset /cellBin/blockIndex
dataset /cellBin/blockSize
dataset /cellBin/cell
dataset /cellBin/cellBorder
dataset /cellBin/cellExp
dataset /cellBin/cellTypeList
dataset /cellBin/gene
dataset /cellBin/geneExp
}
}
9. 测序饱和度
9.1 测序饱和度文件示例
bash$ cat /path/to/output/08T.saturation/sequence_saturation.tsv
sample bar_x bar_y1 bar_y2 bar_umi bin_x bin_y1 bin_y2 bin_umi
0.05 10161815 0.0526077 1 9627224 10161815 0.0686479 2668 5671
0.1 20323630 0.0976586 1 18338852 20323630 0.116931 4021 10804
0.2 40647260 0.172483 1 33636299 40647260 0.194497 5585 19816
0.3 60970888 0.232096 2 46819785 60970888 0.254876 6499 27583
0.4 81294520 0.281427 2 58416019 81294520 0.30439 7118 34415
0.5 101618144 0.322769 2 68818922 101618144 0.345701 7570 40544
0.6 121941776 0.358552 2 78219368 121941776 0.381207 7926 46083
0.7 142265392 0.389758 2 86816303 142265392 0.412077 8228 51147
0.8 162589040 0.417262 2 94746848 162589040 0.439195 8447 55820
0.9 182912656 0.441618 2 102135187 182912656 0.463109 8659 60173
1 203236286 0.46353 3 109030103 203236286 0.484589 8817 63276
bash
$ cat /path/to/output/08P.saturation/sequence_saturation.tsv
sample bar_x bar_y1 bar_y2 bar_umi bin_x bin_y1 bin_y2 bin_umi
0.05 21388070 0.0631674 1 20037040 21388070 0.0888825 126 10218
0.1 42776140 0.120048 1 37640944 42776140 0.150368 128 19197
0.2 85552280 0.217101 2 66978781 85552280 0.249677 128 34159
0.3 128328424 0.296905 2 90227130 128328424 0.32905 128 46016
0.4 171104560 0.363266 2 108948120 171104560 0.394026 128 55564
0.5 213880704 0.418843 3 124298217 213880704 0.447933 128 63392
0.6 256656848 0.466254 3 136989686 256656848 0.493682 128 69865
0.7 299432992 0.506823 3 147673538 299432992 0.532617 128 75314
0.8 342209120 0.541944 3 156750875 342209120 0.566236 128 79943
0.9 384985248 0.572588 3 164547428 384985248 0.595516 128 83919
1 427761398 0.59948 3 171326815 427761398 0.62115 128 86073
10. report-PT
10.1 分析结果统计报告示例
$ head /path/to/10.report/A02677B5.statistics.json"
{ "version": "version_v2",
"1.Filter_and_Map": {
"1.1.Adapter_Filter": [
{
"Sample_id": "V350248064_L03_read_1",
"getCIDPositionMap_uniqCIDTypes": "551164442",
"total_reads": "336354182",
"CID_withN_reads": "523 (0.00 %)",
"mapped_reads": "269471919 (80.12 %)",
10.2 蛋白组和转录组分析结果统计报告示例
分析结果统计报告为HTML格式,可通过SAW Github获取。报告静态示例可参考文件《SAW v7.1 pt report》。




